Perlara’s high throughput screen progress in Mucolipidosis IV (MLIV) flies…
In this blog post, I’ll tell you about Perlara’s high throughput screen progress in MLIV flies. Last year, Tom Hartl reviewed some details of MLIV disease biology and described how worms and flies could be used in a drug discovery screen in his blog post “A case for Using Model Organisms in MLIV drug discovery”. If you remember, MLIV is a rare lysosomal storage disease with an early onset in patients that is caused by mutations in the gene MCOLN/TRPML1. A mutation on the TRPML1 gene can lead to a malfunction on the protein that is controlled by the gene. This protein malfunctioning affects the proper formation of lysosomes inside of the cell that are no longer marked to be move out of the cell membrane since they are not yet a full lysosome. An increase number of unmarked lysosomes inside of the cell membrane will lead to a decrease in the lysosome pH which in turns affect other processes that happen inside of the cell such as the regulation of autophagy by which macromolecules and organelles are repurpose during time of starvation. In the absence of Trpml1 functioning gene there is an accumulation of autophagy substrates.
Making TRPML1 mutant flies
The first step toward a high throughput screen was making a fly with a mutation in the fly ortholog of MCOLN/TRPML1 called trpml. In a study from 2008, Kartik Venkatachalam from Craig Montell’s lab at Johns Hopkins introduced mutations into the fly trpml gene. Much like patients affected by MLIV, Kartik’s trpml fly mutant flies had locomotor defects, progressive neurodegeneration, and early lethality (during the flies pupal stage). With the help of GenetiVision, we generated our own trpml mutant fly line by using the CRISPR/Cas9 gene editing system (you’ve probably heard much about CRISPR in the news, lately). We replaced the region of DNA coding for trpml with DNA coding for green fluorescent protein (GFP). This mutant (trpml∆) is a null for trpml since most of the coding region is delete (Figure 1.2).

Figure 1.2: A) Location of the excisions made into the trpml gene by Kartik Venkatachalam in 2008 to generate two mutant fly lines. B) Location of our 2kb deletion into the trpml gene using CRISPR/Cas9 for the Perlara’s fly null mutant
Characterizing mutant flies
Now that we had our trpml∆ mutant flies, we moved to characterize them. Nearly all of our trpml∆ flies survived through their larval stage, but died during the pupal stage before emerging as adults (Figure 1.3), just like Kartik had observed with his trpml mutants. Also to confirm that the pupal lethality was caused by our trpml∆ mutation, we ectopically drove the expression of trpml in trpml∆ mutants. We observed rescue of pupal lethality phenotype with expression of this transgene, which suggested that the observed phenotype was specific to our null mutation. Most or all of the lethality we observed in trpml∆ flies took place during the pupal stage suggesting that we could potentially conduct a drug treatment prier to pupal stage to rescue this phenotype.

Figure 1.3: We observe normal development to pupal stage on both Trpml∆ heterozygotes and Trpml∆ homozygotes (Blue and Yellow bars); however, only the Trpml∆ heterozygotes animals will develop into full adults (Green bar) and 0% of Trpml∆ homozygotes will become adults.
Developing a Screenotype
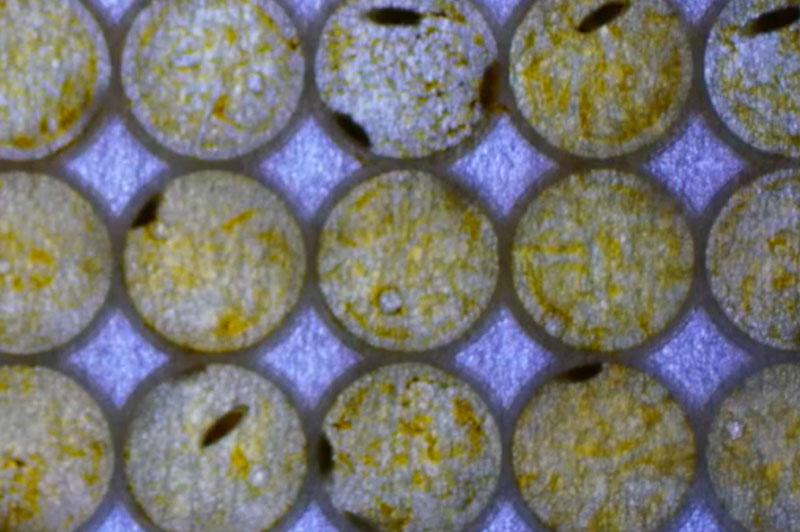
Now we needed to turn this phenotype into a screenotype that could be used for a high throughput screen. To do this, we first determined that we could culture flies in a 96-well plate from 1st instar larvae to fully developed adults without introducing any stress into the animal. Next, we needed to determine if we could obtain quantifiable data in a high throughput manner using 96-well plates and with the help of Modular Science we adapted our larval size fly imager software to register motion. Since our trpml∆ flies are 100% pupal lethal, a small molecule that rescues this phenotype would mean that we would see adult moving flies; movement that is detected and quantified with the fly imager (Figure 1.4 B). The final step to determine if we had an optimal screenotype was to determine a strong dynamic range between our negative and positive controls. Our negative control group was trpml∆ homozygotes in vehicle only (this are 100% pupal lethal) and since MLIV has a recessive pattern of inheritance, our trpml∆ heterozygote sibling’s flies fully developed into adult flies and could be used as our positive control. Figure 1.4 A) is a 96-well plate with trpml∆ heterozygotes on the top four rows (positive control) and trpml∆ homozygotes on the bottom four rows (negative control)
A)
B)

Figure 1.4: A) a video of trpml∆ heterozygotes animals on top four rows where we observe adult fly movement, and trpml∆ homozygotes animals on the bottom four rows where we do not observe adult fly movement. B) Shows quantifiable fly imager data of our fly imager with trpml∆ heterozygotes and trpml∆ homozygotes.
Validating the Screen
Now that we had a reliable and quantifiable screenotype we were ready to start a small molecule high throughput drug screen. On each 96-well pate format we test 80 compounds, 8 positive, and 8 negative internal controls per plate (Figure 1.6). We conducted a pilot screen of 2,532 compounds known compounds (~600 compounds FDA approved drugs, 800 compounds have reached clinical trail stages in USA, 400compounds have been marketed in Europe and Japan, 600 compounds are tool compounds with known bioactivities, and 800 compounds are natural products) in 3 replicates in a period of 2 months obtaining 7 compounds that rescued our pupal lethality phenotype across the 3 replicates (Table 1). We are currently passing those 7 compounds through a secondary screen in vials with a larger sample size to determine the most potent compound that rescues our trpml∆ pupal lethality phenotype.

Figure 1.6: Plate format of pilot high throughput screen. Each testing plate had 8 positive control wells and 8 negative control wells and 80 testing compounds per plate.
| Molecule Name | Plate Name | Phenotype |
|---|---|---|
| PLs-XXXXX16 | PL0158.3 | Adults moving in the well in 3 replicates |
| PLs-XXXXX56 | PL0159.4 | Adults moving in the well in 3 replicates |
| PLs-XXXXX21 | PL0160.2 | Adults moving in the well in 3 replicates |
| PLs-XXXXX13 | PL0161.1 | Adults moving in the well in 3 replicates |
| PLs-XXXXX80 | PL0163.3 | Adults moving in the well in 3 replicates |
| PLs-XXXXX43 | PL0164.2 | Adults moving in the well in 3 replicates |
| PLs-XXXXX58 | PL0164.4 | Adults moving in the well in 3 replicates |
Table 1: List of 7 compounds that showed rescued across the 3 replicates of the pilot screen conducted with trpml∆ mutants.
To summarize, we found that our trpml∆ mutant flies die early in life during the pupal stage. We developed a high throughput screen to rapidly score this phenotype. We ran a pilot screen and found seven chemical compounds that partially rescued the pupal lethality in these mutants, which demonstrates, that we have a reliable screenotype. Now we’re ready to embark ourselves into screening a much larger library of novel chemical compounds with confidence that we will see an effect if they can rescue the disease phenotype. Finally, the worm and cell team at Perlara have already started screening their respective MLIV mutants too, so we’ll soon have results to compare across the three models. Stay tuned for more MLIV findings!.