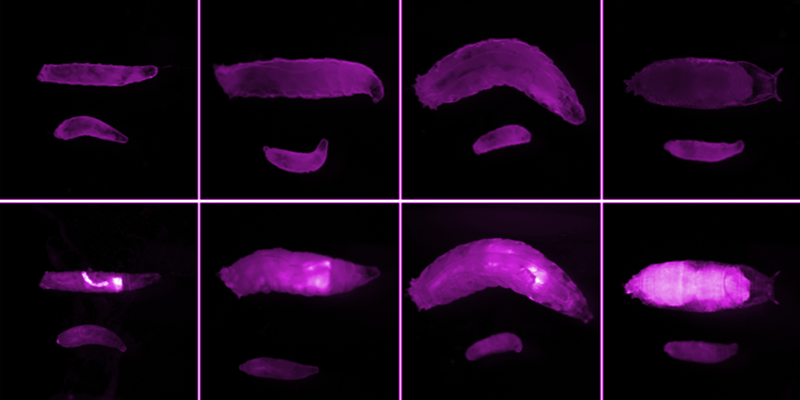
We are currently carrying out screens to find small molecules that could one day become a Niemann-Pick Type C (NPC) therapy. Patients with NPC carry mutant, or defective, copies of the npc1 gene. Like many human disease genes, the npc1 gene also exists in fruit flies. Flies carrying mutant npc1 genes do not really develop into flies. They arrest at an early larval stage because they are deficient in a steroid hormone that promotes developmental progression (Fig. 1A). It is likely that most of the cellular defects in npc1 mutant flies also exist in NPC patient cells. Therefore, we are using npc1 mutant flies to search for potential NPC therapies. The experiment is straightforward: npc1 mutant larvae are fed a drug-like compound, and then over the course of a week, we observe whether the larvae develop beyond the first larval stage. In January and the first half of February, we did this experiment with 10,480 different compounds.

I mentioned that 100% of npc1 mutant larvae arrest at the larval stage and therefore do not become flies. Flies, not larvae, are capable of reproduction. So, we culture flies that carry one mutant npc1 gene, and one functional npc1 gene. These heterozygotes can breed with one another, and 25% of their progeny carry two bad npc1 genes (Box 1). It’s these homozygotes we want to collect for drug screening. However, the young npc1 homozygous larvae have no overt phenotype. Thanks to a nifty genetic tool called a balancer chromosome, the heterozygotes do. The heterozygotes express the green fluorescent protein (GFP) on this balancer, and therefore are bright green (Fig. 1B, Box 1). The desired homozygotes can be identified by the absence of GFP.
 This Punnett Square indicates the genotypes and phenotypes of progeny from a cross between two npc1 heterozygotes, or carriers. Half the gametes from npc1 heterozygous females and males (eggs or sperm) will carry a chromosome with an npc1 mutation. The other half of their gametes will carry the “CyO, GFP” chromosome. Therefore, when flies of the npc1/CyO, GFP genotype mate with one another, 25% of their offspring will be doubly mutant for npc1 (top left quadrant), 50% will be npc1/CyO, GFP (top right and bottom left quadrants), and 25% will be CyO, GFP/CyO, GFP. CyO, GFP/CyO, GFP offspring are embryonic lethal, so do not survive to become larvae. In early stage larvae, the ratio of npc1/ npc1 to npc1/CyO, GFP larvae is 1:2.
This Punnett Square indicates the genotypes and phenotypes of progeny from a cross between two npc1 heterozygotes, or carriers. Half the gametes from npc1 heterozygous females and males (eggs or sperm) will carry a chromosome with an npc1 mutation. The other half of their gametes will carry the “CyO, GFP” chromosome. Therefore, when flies of the npc1/CyO, GFP genotype mate with one another, 25% of their offspring will be doubly mutant for npc1 (top left quadrant), 50% will be npc1/CyO, GFP (top right and bottom left quadrants), and 25% will be CyO, GFP/CyO, GFP. CyO, GFP/CyO, GFP offspring are embryonic lethal, so do not survive to become larvae. In early stage larvae, the ratio of npc1/ npc1 to npc1/CyO, GFP larvae is 1:2.
The development of a high-throughput screen with fly larvae posed several challenges. Fly culturing had to be miniaturized so that experiments would fit into a manageable space and would use a small amount of the costly compounds from our library. We developed a food recipe that supports larval development at a low volume. We also learned to raise a great quantity of flies to collect hundreds of thousands of fly embryos daily. That work was greatly supported by advice from Rebeca Choy, who cultured thousands of flies while working with David Agard at UCSF. All in all, our biggest challenge was mastering a method to rapidly identify and then quickly transfer npc1 mutant larvae into compound-laced food in wells of microtiter plates. This was necessary to quickly screen thousands of drug-like compounds.
To expedite npc1 mutant larvae identification and transfer, we utilize Union Biometrica’s Biosorter. Briefly, in preparation for the Biosorter, a mixture of npc1 heterozygous and homozygous larvae are submerged in a liquid. Then, the larvae are drawn through a capillary where one by one they pass in front of a laser. The amount of laser light that passes through and around the larvae is detected by the instrument. A smaller larva will allow more light to pass then a larger larva will. Thus, a relative size measurement for each larva is created called “extinction.” The laser can also excite GFP, and the amount of light emitted from GFP is detected and measured. This data collected with the Biosorter can identify larvae through their size, and determine whether they are GFP positive or negative. In Figure 2A, plotted on the X-axis is the amount of GFP detected. As expected, there are two populations. The homozygotes are the cluster on the left, and the heterozygotes are the cluster on the right. Plotted on the Y-axis is an indication of larval size based on the amount of light that passed through them, called “extinction”. Large larvae are higher on the Y-axis. There is a range of larval sizes, probably because these larvae range from 0 to 6 hours old, and larvae grow with age.
There are some interesting parts of Figure 2A to note. First, notice that there are ~10,000 homozygotes and ~20,000 heterozygotes in the population. This 1:2 ratio is because another population, the CyO/CyO progeny, do not develop beyond embryonic stages and therefore don’t become larvae (Box 1). Second, look at the heterozygous population of larvae in Figure 2B. There are many more large larvae (black dots and blue dots) present that don’t exist in the homozygous population. At that size of larvae, the ratio homozygotes to heterozygotes is >1:10. That’s because the npc1 homozygotes are developmentally arrested. You can also see a slight accumulation of npc1 homozygotes at a size where their ratio to heterozygotes of the same size is greater then 1:2 (** in the graph, dark blue dots). That size must be the point in npc1 homozygous development when their steroid hormone deficiency is first manifested as arrested growth.

Figure 2. npc1 homozygous and heterozygous larvae plotted via the Biosorter. (A) Approximately 30,000 npc1 heterozygous and homozygous larvae were ran through a Biosorter. The X-axis is extinction, or size of the larvae, based on the amount of light that was detected to pass through and around them. The Y-axis is the amount of GFP detected. The amount of GFP detected revealed two populations, GFP negative homozygotes on the left, and GFP positive heterozygotes on the right. As expected, the ratio of homozygotes to heterozygotes is 1:2. (B) Homozygous and heterozygous larvae are binned at 7 different sizes. The overall ratio is 1:2, but becomes 1:11.2 at the largest size (black dots) because few homozygotes grew to that size, and is 1:1.5 at a small size (dark blue dots) because the growth of many npc1 homozygotes was arrested.
The Biosorter can identify npc1 homozygous larvae, but can it transfer them to wells of a microtiter plate? YES! We routinely use the Biosorter to dispense a fixed amount of npc1 homozygotes to micro titer plate wells, where they then start to consume compound-laced food.
The Biosorter is not a routinely used instrument in Drosophila research. In fact, we don’t know of any fly labs that are using this tool. There may be different and interesting applications were it could used to isolate rare individuals from a population. Let us know if you have an interesting application idea for the Biosorter, or if you happen to be using this great tool in Drosophila research.