In late June, I attended the Mucolipidosis Type IV Foundation’s ML4 Family and Research Conference in Atlanta. It was my first time attending and presenting at this intimate, biennial gathering of families, scientists and physician scientists, some of whom have known each other for years before the ML4 gene (MCOLN1) was cloned almost twenty years ago. In this post, I first recap my experience at the ML4 Conference 2018, then provide an overview of Perlara’s work in developing ML4 worm and fly screens.
Honored to be here, and excited to connect and collaborate with families and researchers to advance toward a cure for Mucolipidosis Type IV, or ML4 pic.twitter.com/B9HqrS7lbx
— Ethan Perlstein (@eperlste) June 24, 2018
What is ML4?
All lysosomal diseases are devastating, but ML4 is particularly vicious. Affected children appear healthy at birth, and for the first few months of life. Unless a telltale corneal clouding is evident very early, ML4 kids aren’t diagnosed until they’re referred to a neurologist, and then to a geneticist, because of psychomotor delay and missed developmental milestones. Kids progressively lose the ability to walk on their own and are wheelchair bound; they are nonverbal, and cognitively peak at a 15-18 month old level. They go blind as young teenagers from retinal degeneration, and don’t survive beyond their 20’s and 30’s.
This cruel reality was brought home by the parents of Brian Wien, who passed away from ML4 last year. His yahrzeit was a month before the conference. He would have turned 37 this year, just two years younger than me. His parents eulogized him with poise and dignity. There wasn’t a dry eye in the room as they paid tribute to their son and explained why they decided to give the most selfless gift to the ML4 community: his vital organs were donated for research to the Maryland Brain and Tissue Bank.
 The Wien family, 1990s (photo source)
The Wien family, 1990s (photo source)
Past, present and future of ML4
Even as a relative newcomer to the field, I could immediately tell that deep professional and personal attachments had been forged between ML4 families and (almost exclusively academic) researchers over decades. In the 1990’s, the collective energy was focused on a mad dash to clone the gene. In the 2000’s, the focus was on developing animal and cellular models. In this decade – especially in the last three years – the tide has decidedly turned toward translational research, and it’s early days still.
As the only person in attendance at ML4 Conference 2018 from a biotech company working on ML4, I felt honored to be invited into this community. As often happens in the rare disease world, I was there because of the persistence of one person: the ML4 Foundation Director Rebecca Oberman. I was first connected to Rebecca by an email from Kartik Venkatachalam in December 2015. Kartik is a professor at University of Texas, and first author of the 2008 paper describing the ML4 fly model. I met Rebecca in person at WORLD (We’re Organizing Research on Lysosomal Diseases) in February 2016, and told her that ML4 was the second-most de-risked program from a model organism perspective (after Niemann-Pick C/NPC, our lead program).

ML4 Foundation Executive Director Rebecca Oberman with Eden Gold (photo source)
In the summer of 2016, Tom Hartl (now at BioMarin) sketched out our plans to use worm and fly models of ML4 for drug discovery as part of a lysosomal diseases collaboration with Novartis that was, at the time, still in contract negotiation. ML4 was on our radar as early as 2014, because the foundational studies in worms and flies had been published in 2005 and 2008, respectively. Last year, we developed ML4 worm and fly (and ML4 patient fibroblast) high-throughput assays, and piloted them in the MicroSource Spectrum collection.
(video source: www.ml4.org)
I came to Atlanta earlier this summer to break bread with ML4 families, to learn from ML4 researchers, and to share preliminary screening results from our multi-species ML4 screening campaign, using a curated bioactive collection (“MoA Box”) that Novartis has run extensively through cell-based screens but not whole-animal screens. We’re currently validating hits, and soon we’ll start writing up this work as a proof-of-concept publication for model organism-based drug discovery. We hope to advance towards an unbiased high-throughput drug screening campaign for novel chemical entities with the ML4 Foundation as a PerlQuest partner.
Scientific highlights of ML4 Conference 2018
Before I review our data, I want to relay synopses of all the outstanding research presented over two intense days of wall-to-wall talks. The first day of the ML4 conference 2018 was actually devoted to family support, including the aforementioned remembrance by Mike and Nanette Wien, and a session on how to fundraise, which I’ve never seen at a rare disease conference before. The conference emcee was ML4 Foundation Board President Randy Gold who, with his wife Caroline, took the leadership reins a decade ago, when the torch was passed from the older generation of ML4 families to the younger generation. The Golds, and their daughter Eden, were profiled in 2013 and again last year.
Pediatric neurologist Albert Misko kicked off the meeting with updates on the planned ML4 natural history study that will take place at Massachusetts General Hospital. Misko mentioned the NPC Clinical Severity Score as a model for how the ML4 natural history study should be conducted. Throughout the meeting, Albert ran a clinic for the kids, collecting valuable data and biological samples. Next up was Sue Slaugenhaupt, the geneticist from MGH who led the effort to clone the ML4 gene when she was at NIH. In my experience, it’s all too rare that the geneticist who cloned a rare disease gene is still active in the field, training the next generation of ML4 researchers, integral to the creation of mouse models of the disease, and a genuinely nice and collaborative person.

To wit, Yulia Grishchuk trained with Sue. Yulia presented data on the ML4 knockout (KO) mouse in collaboration with Levi Wood’s and Kirill Kiselyov’s labs. The ML4 KO mouse starts to decline around two months of age, and all KO mice are dead by nine months. Early in the disease progression, Yulia’s lab found that microglia and astrocytes are activated, and pro-inflammatory cytokine levels are elevated, both in vivo and in vitro, using a primary astrocyte culture system. Arguably the most exciting results of the meeting sprang from these studies. Levi Wood presented data that an FDA-approved immunomodulator for multiple sclerosis, called fingolimod, dissipates the cytokine storm and restores lysosomal homeostasis. Mouse validation studies are currently underway. A paper, unfortunately paywalled, details the science behind this progress, and a story appeared in the Atlanta Journal Constitution about it.
Perlara’s ML4 worm screen
The worm and fly team will follow up on this post in the fall with deeper scientific dives, but for now I want to share at a high level the preliminary results of our ML4 worm and fly drug screens. First up, the ML4 worm screen. The Fares Lab at the University of Arizona developed multiple ML4 models in worms, whose MCOLN1/TRPML1 ortholog is called CUP-5. The Fares Lab described two cup-5 mutants: a null mutant that exhibits early lethality and so is unsuitable for a high-throughput drug screening campaign; a hypomorphic mutant (ar465) with a missense mutation in a conserved residue (G401E) that is perfectly viable and develops apace, but has specific cellular defects that are analogous to what is observed in ML4 patient fibroblasts.
What’s unique about this hypomorph model is that green fluorescent protein (GFP) is expressed under a muscle-cell promoter (Pmyo-3::ssGFP) and released into the body cavity of worms, where it is rapidly taken up and degraded by phagocyte-like cells called coelomocytes. The coelomocytes in ar465 mutant worms are unable to digest the GFP they take up from the body cavity, and so they accumulate GFP as though it were a storage material. As shown below, we can detect and measure the intensity of GFP accumulation in coelomocytes in live (but immobilized) ar465 mutant worms, and screen for compounds that reverse or prevent GFP accumulation in coelomocytes without affecting growth, development or fecundity.
 Wildtype CUP-5 worms on the left; cup-5 mutant worms on the right
Wildtype CUP-5 worms on the left; cup-5 mutant worms on the right
Here’s the 30,000 foot view of the MoA Box screening results, showing robust separation of negative controls (untreated cup-5 mutant worms) and positive controls (untreated wildtype CUP-5 worms), as well as a random distribution of hits, depicted as asterisks, across the library. We screened a total of four independent replicates (reps 1-4). Across all four replicates, 57 compounds were hits, of which 28 were true hits (as categorized below) and 29 were toxic.

When we examined the hit wells closely, we were pleasantly surprised to observe three distinct classes of hit compounds, as shown below.

The first class we call GFP reduction, which means coelomocytes still accumulated GFP, but to a lesser extent. The second class we call GFP clearance, which means we didn’t detect much or any GFP accumulation in coelomocytes. The third class we call GFP diffusion, which means the overall GFP fluorescence intensity is unchanged, but the signal is spread throughout the body cavity instead of concentrated in coelomocytes. The first two classes are the easiest to interpret, especially when the reduction or clearance of GFP in coelomocytes is associated with reduced growth and slower development, i.e., non-specifically toxic compounds. The third class – GFP diffusion – may speak to a different mechanism of action wherein GFP is never taken up by coelomocytes in the first place, or, potentially more interestingly, wherein accumulated GFP is expelled from coelomocytes, e.g., lysosomal exocytosis.
Perlara’s ML4 fly screen
Next up, the fly model. As I mentioned above, the ML4 fly model was developed by Kartik Venkatachalam when he was a trainee in Craig Montell’s lab. We generated fresh trpml-/- genetic null mutant flies by creating a 2kb deletion spanning exons 1-3. Like Kartik showed in his 2008 paper, our null trpml-/- mutant flies exhibit pupal lethality. So we designed a high-throughput eclosion assay in 96-well plate format that allowed us to detect the presence or absence of living adult flies in a custom imager. Here’s an example of what a representative plate looks like from the MoA Box screen. The leftmost column contains the negative controls (trpml-/- flies) and the rightmost column contains the positive controls (trpml+/- flies). Can you spot the two wells with moving flies?
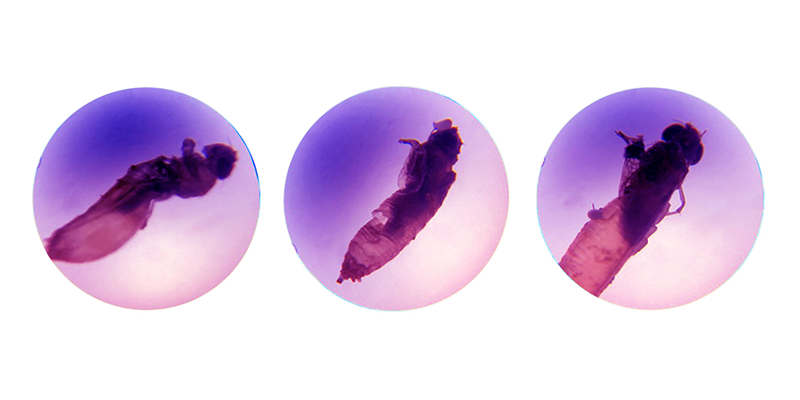
Activity/movement is a very stringent readout. After we recorded each plate, the fly team manually inspected each well looking for eclosed but dead adult flies – or what are called “half-out” flies – which complete metamorphosis but die as they are emerging from the pupal case, as shown here:

As in the ML4 worm screen, we observed complete separation of positive and negative controls in the activity readout. The manual score readout is less stringent, so we see a modicum of overlap between positive and negative controls because there is a low but consistent rate of half-out trpml-/- flies (but never live adult trpml-/- flies).

The hit overlap between the three replicates using the activity readout (top) and the manual score readout (bottom) reveal that while no compounds were activity hits in 3/3 replicates, three compounds were manual score hits in 3/3 replicates.

Finally, here are the replicate overlaps depicted as a Venn diagram. We’re feverishly analyzing this dataset to understand the overlap across ML4 worm, ML4 fly and ML4 fibroblast MoA Box screens.

Next year, a reunion
The ML4 Conference 2018 concluded with a brainstorming wrap-up session. Gene therapy was discussed. Small molecule therapeutics as well. Yet the irony of it all is that the whole point of this conference – and every other rare disease meeting – is to make the conference unnecessary. Or, as a parent advocate once quipped to me “to turn it from a conference into a reunion.” Amen to that!
Stay tuned for updates in the Fall!