
by Jessica Lao | Nov 19, 2018 | PMM2-CDG |
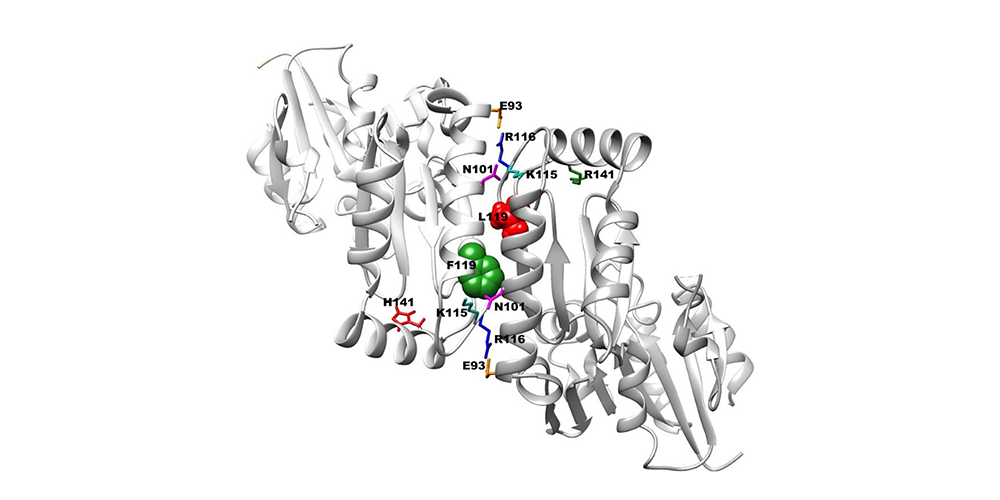
This is a continuation of a previous post, which covered Figures 1 – 4 of Perlara’s preprint on bioRxiv, describing our work in modeling PMM2 patient alleles. This work is currently under review at the Genetics Society of America’s peer-reviewed...

by Jessica Lao | Nov 19, 2018 | PMM2-CDG |
Last September, we submitted Perlara’s second preprint to bioRxiv describing our yeast models of PMM2 deficiency. This work is currently under review at the Genetics Society of America’s peer-reviewed journal G3. Below is the description of the figures from the...
by Ethan Perlstein | Sep 20, 2018 | PMM2-CDG |
Last week, we posted a preprint describing new yeast models of Phosphomannomutase 2 Deficiency, or PMM2-CDG, the most common congenital disorder of glycosylation in the world. This PMM2 model preprint is our second preprint on bioRxiv which, for the uninitiated, is...

by Julide Bilen | Jun 7, 2018 | PMM2-CDG |
An update on the development of PMM2 Drosophila models, and our work to design high throughput drug screen for PMM2 deficiency — Phosphomannomutase 2 deficiency (PMM2-CDG) is the most common type of congenital disorder of glycosylation resulting from a defect in...
by Jessica Lao | May 8, 2018 | PMM2-CDG |
Drug repurposing screen in haploid PMM2-CDG yeast models During Stage 1 of our PerlQuest partnership with Maggie’s Cure, we generated haploid PMM2-CDG yeast models of three pathogenic variants: F119L, R141H, and V231M. We expressed these variants in the yeast...
by Sangeetha Iyer | Feb 20, 2018 | PMM2-CDG |
Perlara and Maggie’s Cure have started a PerlQuest for a glycosylation disorder called PMM-2 deficiency disease – one of the most common congenital disorders of glycosylation. Over the past year at Perlara, we have modeled the pmm-2 deficiency disorder in yeast,...








