Scalable drug discovery platform
Perlara uses a platform of genetically engineered animals (yeast, nematodes, fruit flies and zebrafish) in phenotypic screens to identify orphan drug candidates that reverse disease much faster and cheaper than current approaches. Our proof-of-concept diseases are Niemann-Pick type C (NPC), a lysosomal storage disorder first described nearly a century ago, and NGLY1 Deficiency, a metabolic disease related to proteasome-mediated degradation.
PerlArk™ Platform Science
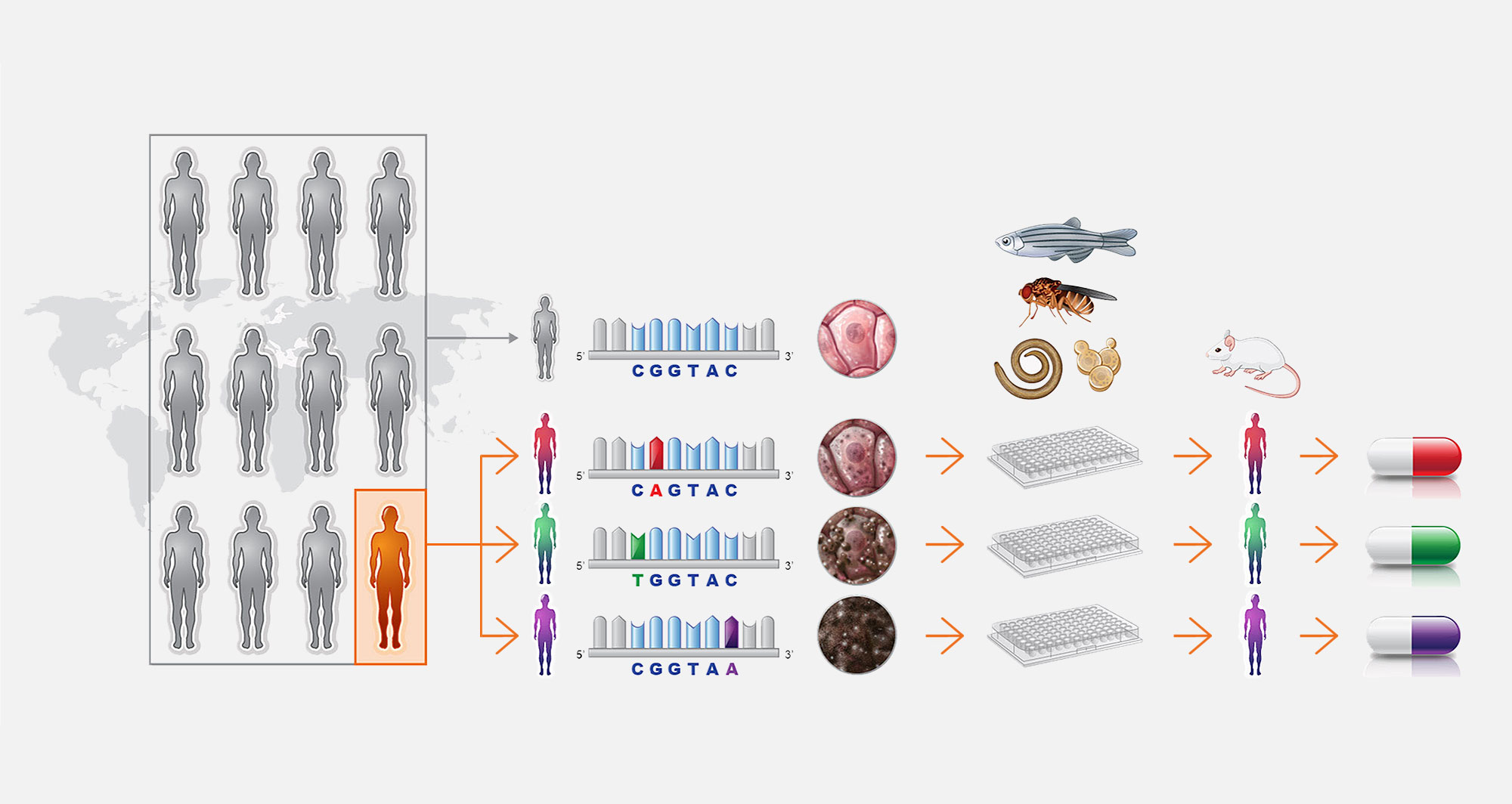
1. What does Perlara do?
Perlara develops precision drug discovery programs for rare genetic diseases. We engineer model organisms to express patient mutations for use in industrial-scale, organism-based phenotypic screens,
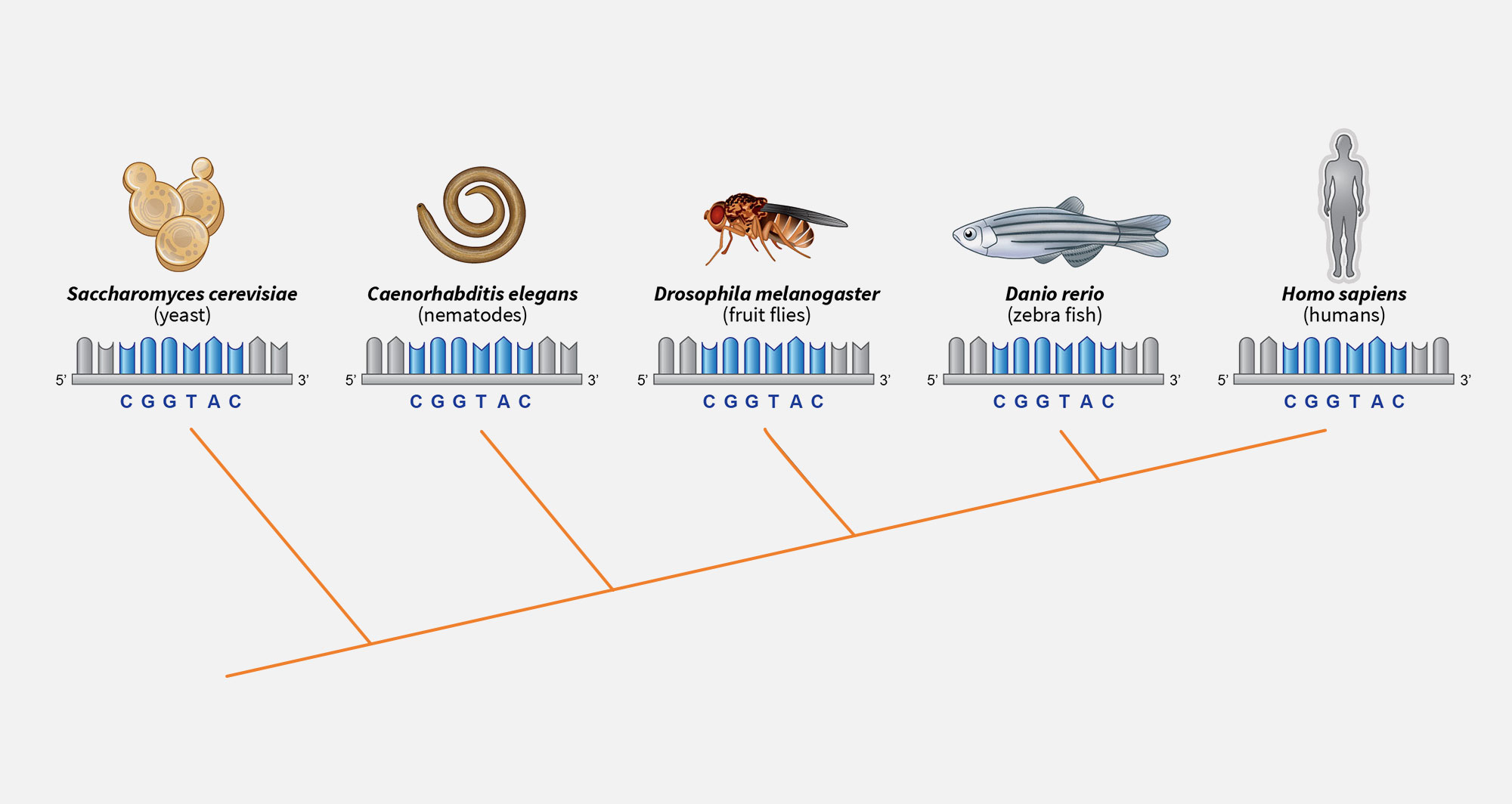
2. Shared genes
All animals sit on the same branch of the evolutionary tree and share thousands of genes. Many of these ancient genes are unchanged at the DNA level despite a billion years of evolution.
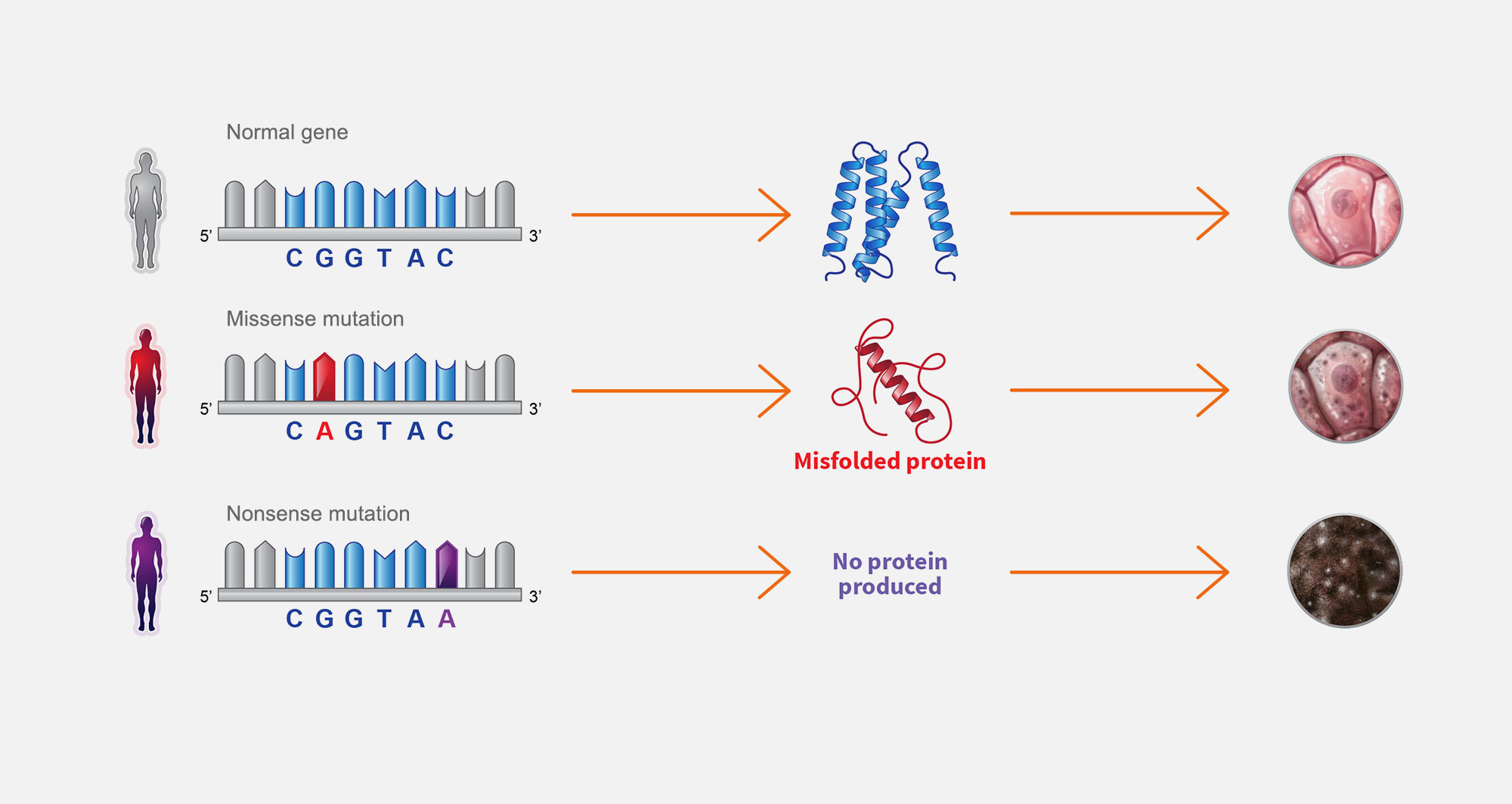
3. Single gene diseases
Single gene diseases are caused by a spectrum of mutations and result in cellular defects. Missense mutations prevent proteins from folding correctly, which causes partial loss of gene function. Nonsense mutations prevent protein production, which a causes complete loss of gene function.
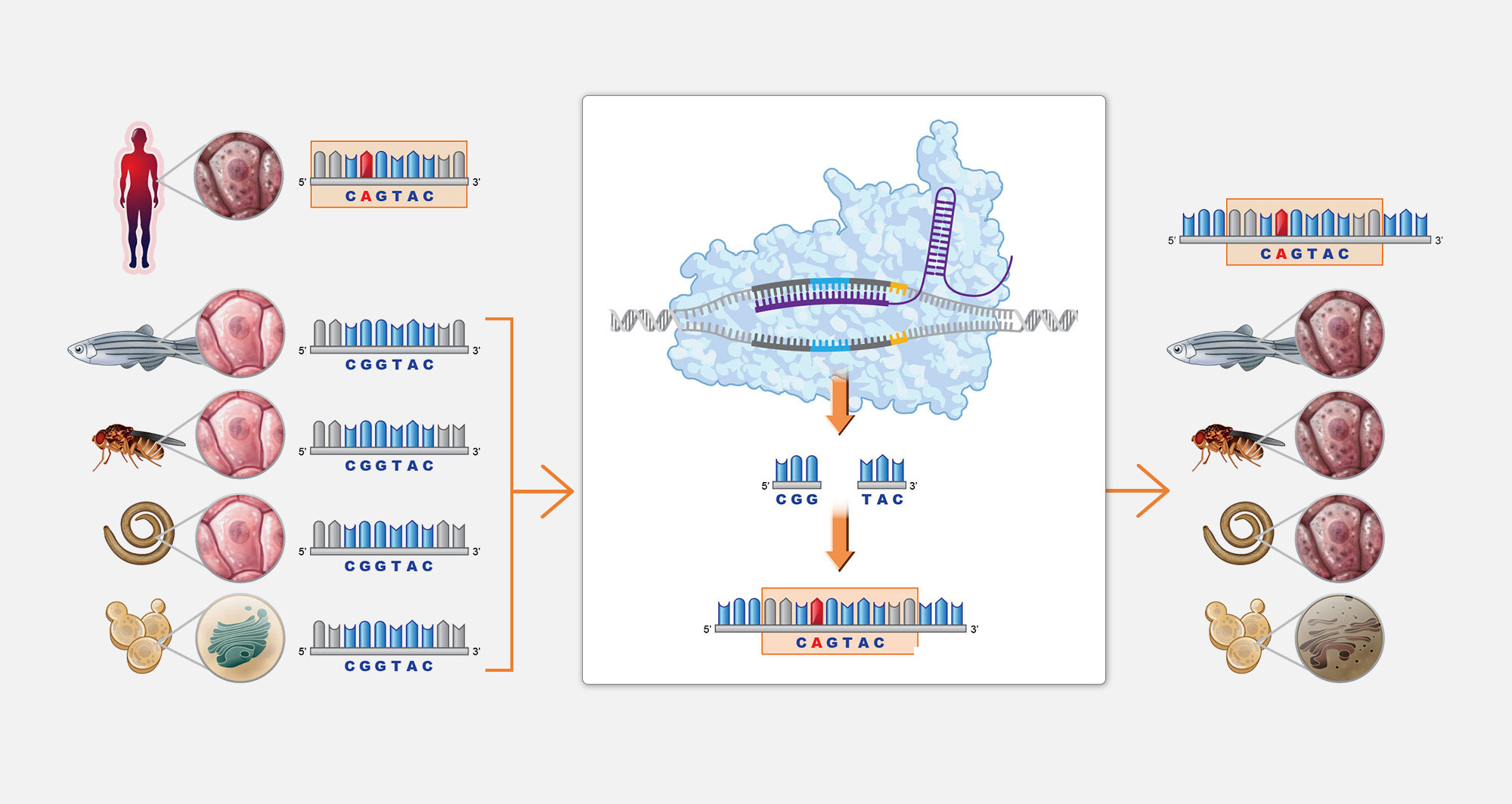
4. Gene editing
Gene editing technology enables us to generate patient-mutation-matched Perlara disease models that leave no mutation behind.
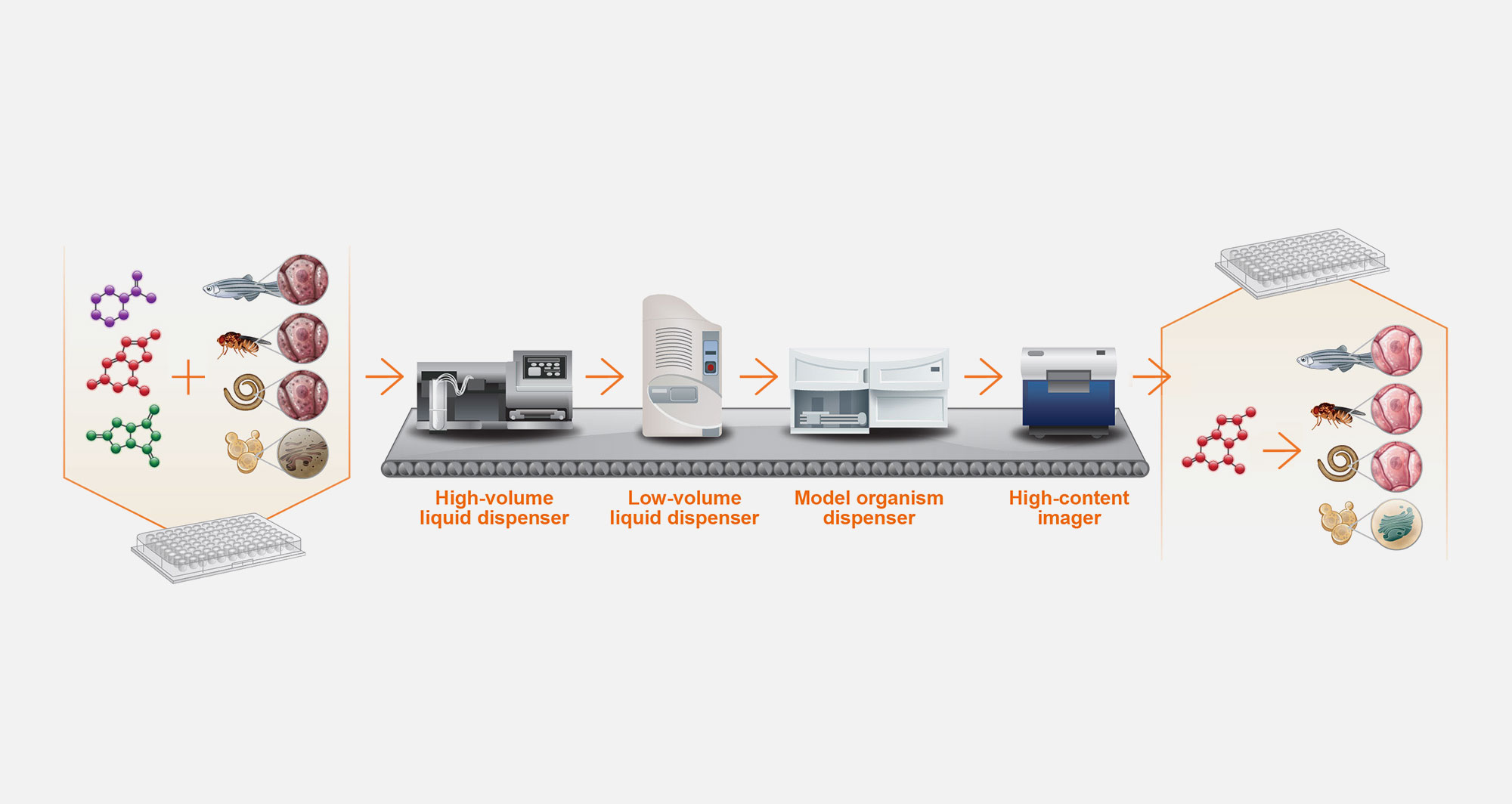
5. Automated drug screening
The resulting disease models manifest cellular defects that can be reversed by novel chemical entities in drug screens. The first version of the PerlArk™ Platform leverages multi-well plate technology and automation.
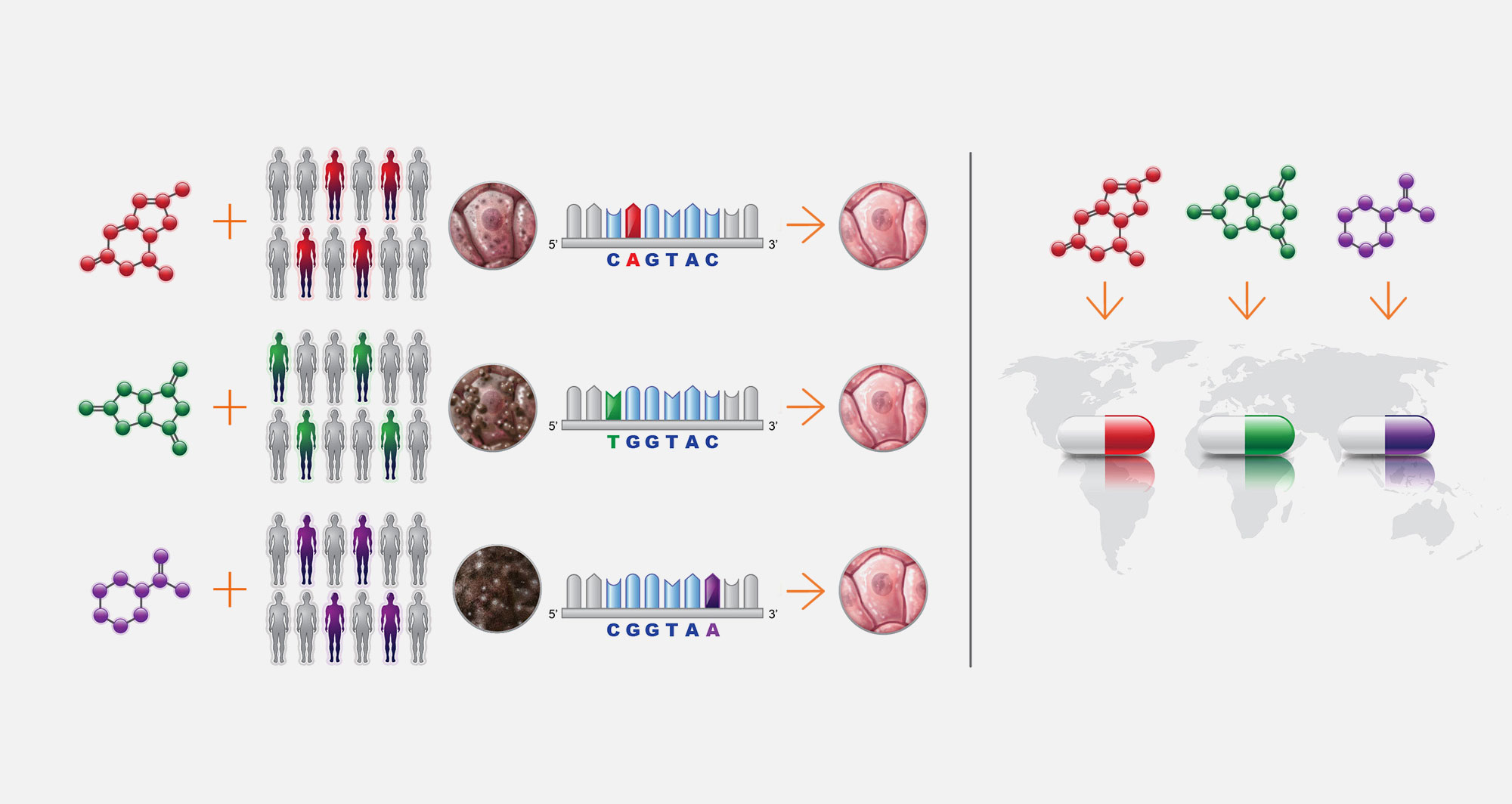
6. Bio-Pharma cure pipeline
We partner with BioPharma and Patient advocacy Groups to advance precision drug candidates to the clinic. Our pipeline begins with childhood single-gene disease involving loss of an ancient cellular process: lipid storage diseases, congenital disorders of glycosylations, peroxisomal biogenesis disorders, mitochondrial diseases, ciliopathies and laminopathies.
Science updates from our Blog
Conducting a GNAO1 yeast screen
by Gabriela Colmenares | December 20, 2018 | GNAO1 | 0 Comments
Modeling Pompe disease in flies
by Madeleine Prangley | December 14, 2018 | GSD II | 0 Comments
Developing an NPA patient fibroblast screen
by Aras Rezvanian | November 30, 2018 | Niemann-Pick Type A | 0 Comments
PMM2-CDG preprint figures – part 2
by Jessica Lao | November 19, 2018 | PMM2-CDG | 0 Comments
PMM2-CDG preprint figures – part 1
by Jessica Lao | November 19, 2018 | PMM2-CDG | 0 Comments
2018 Global Genes Summit recap
by Feba Sam | November 15, 2018 | Rare Diseases | 0 Comments
Grace Science Foundation’s 2018 NGLY1 Conference recap
by Ethan Perlstein | October 25, 2018 | NGLY1 | 0 Comments
2018 Yeast Genetics Meeting recap
by Jessica Lao | October 19, 2018 | Rare Diseases | 0 Comments
Launching the Cori disease PerlQuest
by Nina DiPrimio | October 17, 2018 | GSD III | 0 Comments
Announcing the Sanfilippo syndrome PerlQuest
by Nina DiPrimio | October 11, 2018 | Sanfilippo syndrome | 0 Comments
CFTR-pancreatitis PerlQuest update
by Jessica Lao | October 5, 2018 | Pancreatitis | 0 Comments
Perlara’s 2018 Global Genes presentation
by Naz Dana | October 4, 2018 | Rare Diseases | 0 Comments

